Molecular Detection of Methicillin Resistance Genes (Mec A; Pvl) in Methicillin Resistant Staphylococcus Aureus Isolates from Federal Medical Centre, Yenagoa, Bayelsa State, Nigeria
Abstract
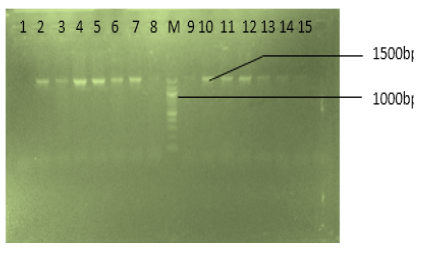
Methicillin resistant Staphylococcus aureus (MRSA) is known to cause serious infections that vary widely in severity and susceptibility to antibiotics treatments. The ability of MRSA to resist specific antibiotics is characterized by its potential to express or harbor certain resistance genes. The mecA codes for methicillin resistance by encoding penicillin binding protein 2A (PBP2A) and reveals low affinity for beta-lactams antibiotics such as methicillin. This study was carried out between February and October 2019, to detect mecA and PVL genes in MRSA from clinical samples collected from Federal Medical Centre, Yenagoa, Diete-Spiff Memorial Hospital and Niger Delta University Teaching Hospital (NDUTH), Okolobiri all in Bayelsa state. A total of 250 clinical specimens {(Urine 136(54.4%), wound 49 (19.6%), endocervical swab 40 (16%) and High Vagina Swab 25 (10%)} collected at random from the 3 hospitals stated above were immediately taken to Niger Delta University, Department of Medical Laboratory Science Research Laboratory for cultured using selective and general purpose media based on standard bacteriological techniques. Pure culture was standardized with 0.5 Mac Farland turbidity method of standardization and subjected to antibiotics susceptibility testing by Kirby Buer agar diffusion method with Muller Hinton agar. Genomic DNA was extracted by boiling method. MecA and PVL were amplified in an AB1970 applied biosystem thermal cycler. Of 250 samples collected, a total of 25 (10%) MRSA isolated comprised of 11(44%) from urine 10(40%) from wound, 2(8%) from High vagina swab and 2(8%) from ECS. The highest frequency of MRSA was in urine. The susceptibility pattern of MRSA isolates showed that the organisms was highly resistant to Ampiclox 25(100%), Amoxicillin 21(84%), Streptomycin 19(76%) and was least resistant to Gentamycin 5(20%). Out of the 25 MRSA isolates, none harboured mecA and PVL gene. Statistical analysis of this study reject the null hypothesis at P = 0.05 level of significance. The results obtained showed that the Staphylococcus aureus isolated in this study do not have resistance genes PVL and Mec-A genes, but could have developed another means of resistance ability to these test antibiotics. Further studies on Methicillin Resistant Staphylococcus aureus (MRSA) resistance ability are advised. Continuous monitoring of methicillin resistance pattern of MRSA isolates and the need for high standards of infection control to prevent its transmission and limit its spread is highly recommended.